Date:
Time:
Location:
Event Description:
Nezar Abdennur
Title: Unfolding genome organization interphase
Abstact:
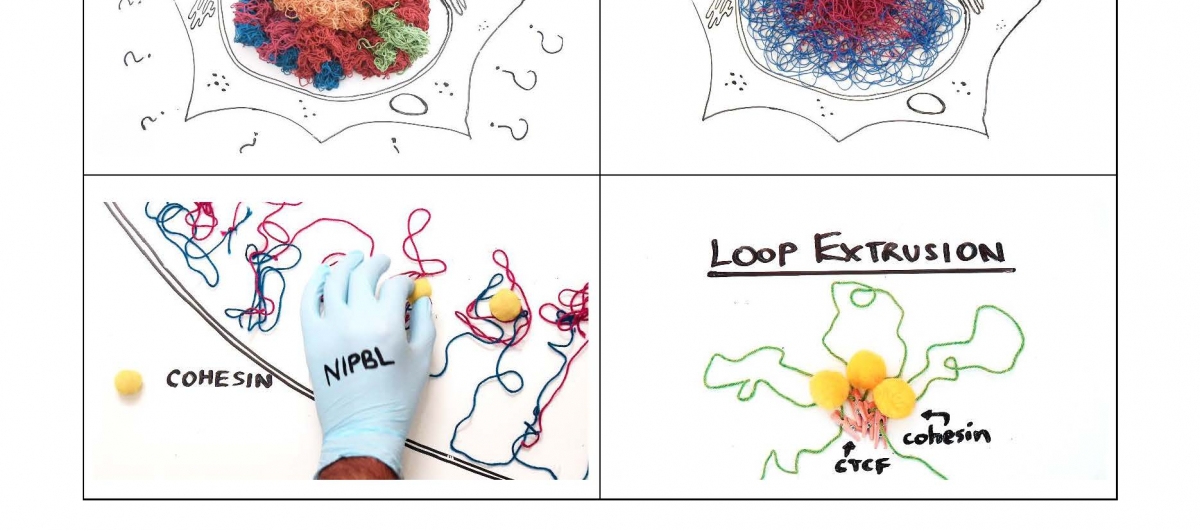
Contact frequency maps from high throughput chromosome conformation capture technologies have revealed several organizing patterns for mammalian interphase chromosomes, including self-interacting topologically associating domains (TADs) which are believed to function as insulating gene regulatory neighborhoods. However, the mechanisms driving these patterns are still unknown. In this thesis, I describe and apply computational methods that test the predictions of a recently proposed loop extrusion model in the context of experimental perturbations of its key molecular players. In the first project I introduce a new data model, file format, and supporting software package to cope with the challenges of the increasing size and resolution of Hi-C datasets, including a parallel and scalable matrix balancing implementation. In the second project, I show that depletion of the Structural Maintenance of Chromosomes (SMC) complex, cohesin, in non-cycling mouse liver cells completely eliminates the appearance of TADs in Hi-C maps while preserving genome compartmentalization. In the third project, I demonstrate that depletion of a closely related SMC complex, condensin II, which plays a major role in mitotic chromosome condensation but is also found in the nucleus in interphase, has no impact on gene expression or the maintenance of genome organization in non-dividing cells. In the final project, I compile further evidence for loop extrusion in interphase by employing a combination of polymer simulations and meta-analysis of several Hi-C studies that performed targeted perturbations to modulate the presence of cohesin and the insulator protein, CTCF, on chromatin. Together, these studies show that mammalian interphase genomes are not folded in a hierarchical fashion, but organized by at least two distinct and antagonistic processes: global compartmental segregation dependent on epigenetic state, and local compaction likely driven by an energy-dependent process of loop extrusion mediated by cohesin complexes and limited by DNA-bound CTCF extrusion barriers.